Download the latest version of DAGchainer from Sourceforge.
The DAGchainer utility consists of a Perl script and a C++ program. Build the C++ program like so:
This will compile the single dagchainer.cpp file, creating a binary with the hostname as the extension. For example, building this on a powermac yeilded the binary dagchainer.powermac
DAGchainer is invoked using the Perl script run_DAG_chainer.pl
The full set of options is revealed with by -h:
############################# Options ###############################
#
## Required:
# -i input file
# input file has format:
# molecule_1 <tab> accession_1 <tab> end5_1 <tab> end3_1 <tab> molecule_2 <tab> accession_2 <tab> end5_2 <tab> end3_2 <tab> P-value
#
#
## DAG chaining scoring parameters:
# -o gap open penalty (default: -0f)
# -e gap extension penalty (default: -3f)
# -g length of a gap in bp (default: 10000) (avg distance expected between two syntenic genes)
# -M Maximum match score (default: 50) otherwise, -log(evalue)
# -Z define constant match score ** use in place of -M
# -D maximum distance allowed between two matches in basepairs. (default: 200000)
#
## Input data filtering:
# -E Maximum E-value (default 1e-5)
#
#
## Output filtering:
# -x Minimum alignment score (alignment pursued until scores fall below this range.)
# default: MIN_ALIGNMENT_SCORE = (int) (MIN_ALIGN_LEN * 2.5 * -GAP_PENALTY)
# -I ignore tandem duplication alignments (overlapping, same mol alignments) (requires -s).
# -A Minium number of Aligned Pairs (default: 6)
# -T only Tandem alignments (implies -s)
#
## Include/Exclude certain molecule comparisons:
# -s include self comparisons.
#
#
## Others:
# -v verbose
# -h print this option menu and quit
#######################################################################
There are many options that can be tuned to preferences based on your application/genome of interest.
Of particular interest is the single input file which contains a list of homologous gene pairs, their genome locations, and their E-value resulting from a BLAST search. The format is described above under the -i option.
DAGchainer includes two large data sets that exemplify its application to both cross-genome synteny discovery and to segmental genome duplication analysis.
Mining Segmental Genome Duplications in Arabidopsis
In order to identify candidate segmental genome duplications in Arabdiopsis by using DAGchainer, we first performed and all-vs-all blastp search using the Arabidopsis protein set. The blastp pairs, the location of each gene, and the corresponding E-value are provided in the sample data file
data_sets/Arabidopsis/Arabidopsis.Release5.matchList
The first few lines of the file are provided below:
1 At1g01010 3760 5630 1 At1g02230 436775 433031 1.3e-45
1 At1g01010 3760 5630 1 At1g02250 439559 437951 1.5e-37
1 At1g01010 3760 5630 3 At3g04420 1172846 1174301 1.9e-33
1 At1g01010 3760 5630 4 At4g01520 659178 656407 5.6e-34
1 At1g01010 3760 5630 4 At4g01540 672629 670483 2.8e-36
1 At1g01010 3760 5630 4 At4g01550 676225 674025 1.5e-44
1 At1g01020 8666 7729 4 At4g01510 642683 644190 2.6e-33
1 At1g01030 12940 11864 2 At2g36080 15158490 15155691 1.2e-39
1 At1g01030 12940 11864 2 At2g46870 19268382 19269314 6.2e-56
Using an accessory script, we removed repetitive matches that contribute noise to the data. This was done clustering all groups of matching genes that fall within 50 kb of each other and reporting only the single highest scoring match in that region. This was done as follows:
../../accessory_scripts/filter_repetitive_matches.pl 50000 < Arabidopsis.Release5.matchList > Arabidopsis.Release5.matchList.filtered
Using this filtered data set, we search for segmental duplications within the Arabidopsis genome by running DAGchainer like so:
../../run_DAG_chainer.pl -i Arabidopsis.Release5.matchList.filtered -s -I
We include the -s option so that chromosome comparisons will be performed against themselves; for example, to identify duplications found when chromosome 1 is compared against itself. The -I is also used so that tandem duplications or artifactual weaved regions are excluded from the output.
The output file is generated with the .aligncoords extension. The output format is identical to the input format with the exception that only those gene pairs found in colinear chains are reported. Each chain is preceded by a header line that indicates the DNA molecules being compared and the number of genes reported within the single chain. For example, the first few lines of our output file are:
% more Arabidopsis.Release5.matchList.filtered.aligncoords
## alignment 1 vs. 1 Alignment #1 score = 8508.9 (num aligned pairs: 188):
1 At1g72300 27224628 27221341 1 At1g17240 5898710 5896521 1.000000e-250 50
1 At1g72330 27237299 27240233 1 At1g17290 5922764 5926086 1.100000e-242 94
1 At1g72350 27243609 27242935 1 At1g17310 5928660 5928007 1.900000e-46 139
1 At1g72420 27266576 27265029 1 At1g17350 5942384 5944265 2.800000e-100 183
1 At1g72450 27279798 27277998 1 At1g17380 5957063 5955647 1.900000e-62 230
1 At1g72490 27294140 27293104 1 At1g17400 5962238 5960939 9.000000e-72 277
1 At1g72520 27312273 27316251 1 At1g17420 5977505 5981377 1.000000e-250 321
1 At1g72620 27344741 27346063 1 At1g17430 5982303 5983912 1.100000e-137 362
1 At1g72630 27348650 27349009 1 At1g17455 5997925 5998269 1.100000e-50 409
1 At1g72650 27353915 27357145 1 At1g17460 5999512 6002539 1.500000e-142 459
...
The inputs and outputs can be examined in the context of a navigable XY plot using the Java XY-plot viewer like so:
% ../../Java_XY_plotter/run_XYplot.pl Arabidopsis.Release5.matchList.filtered Arabidopsis.Release5.matchList.filtered.aligncoords
Inputs to the XY-plotter include the original match list and the output file generated by DAGchainer.
The above command launches the accompanying Java XY-plot viewer and generates a navigable plot for each chromosome comparison, highlighting the diagonals of colinear gene pairs reported by DAGchainer. A screenshot of the display is shown below:

The XY-plotting tool is described in more detail below.
Mining Gene Synteny Among the Tri-Trypanosome Genomes: Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major
To identify syntenic regions among the three tryaponsome genomes, we first searched each protein set among the others to identify candidate homologous gene pairs. The summary output describing the homologous gene pairs found in the context of their corresponding genome locations is provided in the file
/Users/bhaas/CVS/DAGCHAINER/DAGCHAINER/data_sets/Trypanosomes/Tbrucei_vs_Lmajor.match_file
|
Note
|
The T. brucei and L. major genomes are both complete and contiguous. The T. cruzi genome is heavily polymorphic and exists as numerous contigs. Therefore, the number of DNA molecule identifiers for the T. cruzi data is many times greater than that of T. brucei or L. major. |
The genes of these trypansomes lack introns and are separated by short intergenic regions. Instead of describing each gene with a set of genomic coordinates, a simple gene ordering was provided. The complete set of candidate homologous gene pairs is provided at:
data_sets/Trypanosomes/Tbrucei_vs_Lmajor.match_file
The first few lines are as follows:
lma.35_85 lma1.85.m00253 253 253 tba.9_320 tba1.320.m01093 1093 1093 1.3e-29
tba.7_316 tba1.316.m00259 259 259 lma.22_72 lma1.72.m00073 73 73 1.9e-42
lma.18_68 lma1.68.m00064 64 64 tba.8_317 tba1.317.m00080 80 80 4.7e-96
tba.11_322 tba1.322.m00577 577 577 lma.24_74 lma1.74.m00144 144 144 3.2e-16
The first line above indicates that L.major gene lma1.85.m00253 on contig lma.35_85 corresponds to the 253rd gene on that contig. It matches the T.brucei gene tba1.320.m01093 on contig tba.9_320, which is the 1093rd gene on that contig, with a BLASTP E-value of 1.3e-29.
First, we removed the repetitive matches by reporting only the best match within a repetitive cluster of genes found within 5 neighbors along each chromsoome like so:
../../accessory_scripts/filter_repetitive_matches.pl 5 < Tbrucei_vs_Lmajor.match_file > Tbrucei_vs_Lmajor.match_file.filtered
Then, we ran DAGchainer to find syntenic gene pairs like so:
../../run_DAG_chainer.pl -i Tbrucei_vs_Lmajor.match_file.filtered -Z 12 -D 10 -g 1 -A 5
In this case, we used -Z 12 to enforce a constant match score of value 12, required neighboring genes in a single chain to be no more than 10 genes apart (-D 10), set the gap penalty equal to 1 (-g 1), and set the minimum chain length equal to 5 colinear genes (-A 5). The output file was generated with the .aligncoords extension. The first few lines of output are as follows:
## alignment lma.10_60 vs. tba.8_317 Alignment #1 score = 537.0 (num aligned pairs: 53):
lma.10_60 lma1.60.m00057 57 57 tba.8_317 tba1.317.m00393 392 392 1.100000e-13 12
lma.10_60 lma1.60.m00058 58 58 tba.8_317 tba1.317.m00394 393 393 1.200000e-70 24
lma.10_60 lma1.60.m00060 60 60 tba.8_317 tba1.317.m00395 394 394 1.100000e-133 33
lma.10_60 lma1.60.m00062 62 62 tba.8_317 tba1.317.m00396 395 395 7.500000e-101 42
lma.10_60 lma1.60.m00063 63 63 tba.8_317 tba1.317.m00397 396 396 6.500000e-83 54
lma.10_60 lma1.60.m00064 64 64 tba.8_317 tba1.317.m00398 397 397 9.900000e-06 66
lma.10_60 lma1.60.m00065 65 65 tba.8_317 tba1.317.m00399 398 398 1.400000e-149 78
lma.10_60 lma1.60.m00066 66 66 tba.8_317 tba1.317.m00403 402 402 3.000000e-63 81
lma.10_60 lma1.60.m00067 67 67 tba.8_317 tba1.317.m00404 403 403 3.800000e-55 93
lma.10_60 lma1.60.m00072 72 72 tba.8_317 tba1.317.m00414 413 413 1.100000e-78 78
Again, we can use the XY-plotter to examine the matches between genes and those colinear genes reported by DAGchainer.
% ../../Java_XY_plotter/run_XYplot.pl Tbrucei_vs_Lmajor.match_file.filtered Tbrucei_vs_Lmajor.match_file.filtered.aligncoords
This will launch every X-Y plot where matches are reported in the filtered match file. Not all of these will have syntenic regions reported. Accessory tools are available for extracting only regions of interest, as described below.
Here is an example of a region viewed using the XY-plotter that highlights syntenic regions found by DAGchainer between a T. brucei chromosome and a L. major chromosome:
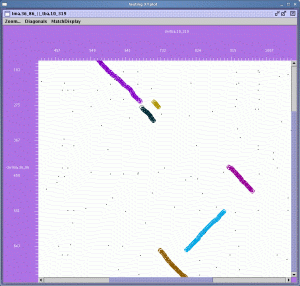
As shown briefly in the examples above, the DAGchainer distribution comes with an XY-plotter tool that can be used to navigate the pairwise matches found using BLASTP in the genome context, and to examine the subset of matches reported by DAGchainer to be syntenic. This tool allows you to zoom in on selected regions and to filter matches based on repetitiveness or by an E-value cutoff. Visit the full DAGchainer XY-plotter tutorial here.
Please reference the following:
Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004 Dec 12;20(18):3643-6. Epub 2004 Jul 9.


