% Java_XY_plotter/run_XYplot.pl data_sets/Arabidopsis/Arabidopsis.Release5.matchList
The plotter can be used with only the initial match list, or with both the match list and the syntenic pairs reported by DAGchainer. First, we highlight the use of the plotter with the match list alone and using the Arabidopsis sample data set.
We can launch the plotter from the root of the installation directory like so:
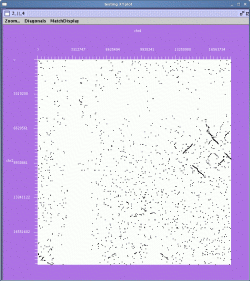
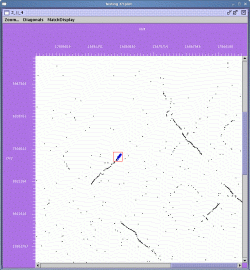
The result is shown below:
In this view, chrosome 2 is ordered along the Y-axis and chromosome 4 is ordered along the X-axis. Each pair of matching genes is shown as a single dot in the plot. The left and right mouse buttons can be used to either select matches of gene pairs or to zoom in on selected regions.
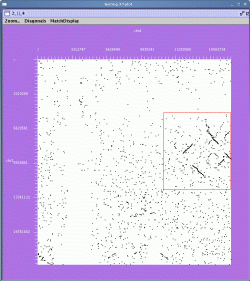
To zoom in on a region, use the right mouse button and drag a rectangle over the region of interest. For example:
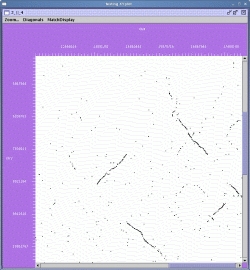
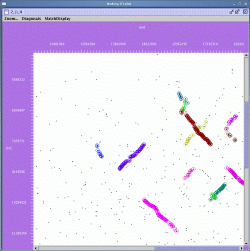
The zoomed display is shown below:
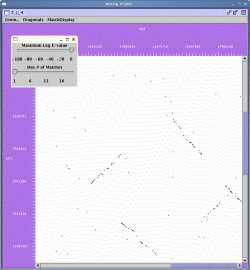
In the zoomed display, the scrollbars at the right and bottom of the display can be used to navigate the region in the viewer.